ubi:argue extraction of arguments and related named entities from biomedical research publications
Researchers of the biomedical domain need to browse the literature in order to find arguments put forth by other researchers regarding the efficiency/toxicity of chemical agents of interest. Browsing through the vast amount of published works can be an intimidating task for humans. Various techniques have been developed which help the researcher in this task, such as indexing, mark-up notes, association via keywords, etc. It is a commonly occurring case however that the source literature comes in an unstructured format, such as PDF, and the researcher needs to search the text of these sources for information.
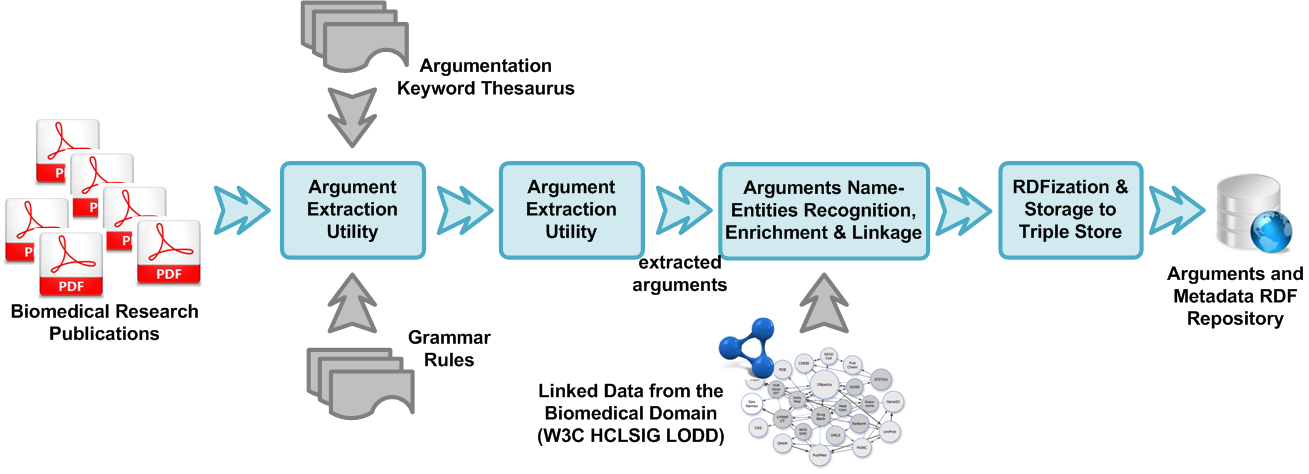
This challenge is addressed by argumentation extraction technology developed by the UBITECH R&D team, called ubi:argue, which tackles the following two problems: (a) the identification of segments of text that correspond to argumentation put forth by the authors of the published work, and (b) the association of each argumentation sentence with the set of named entities (proteins, molecules, active agents, etc.) that are referred to in the sentence.
Argument extraction
Argument extraction takes advantage of the fact that argumentation put forth by scientists in published work, is usually phrased following often used grammatical norms. It is performed by using a set of predefined grammar rules to the text of the publication. Though the rules are not adaptable at runtime, they can easily be modified by an expert user to alter if a different level of sensitivity is required. The rules use the notion of Wordnet’s Synsets, which allows for the formation of thesaurus of semantically linked words and phrases. This can also be modified by the user in case the user wishes to form a different thesaurus for grammar pattern matching.
Extraction of Linked Data
After the argument sentences are extracted, a set of Linked Data instances related to the sentences are identified and retrieved as metadata to each argument. The Linked Data instances matched come from federated SPARQL queries across multiple online sources. All the arguments along with their metadata are converted to RDF triples and stored in an RDF triple store. A researcher can query this data to obtain a vast combination of information that is of interest.
Examples of queries that can be performed can be:
- for a given chemical agent A and a specific disease B, find all the diseases that are referred to the same publication as the afore mentioned agent and disease. The user can thus find candidate diseases for which agent A may have an effect (positive or as a adverse invent) when administered for treating disease B.
- for all the argument sentences found containing reference to a specific hormone receptor, find all the molecules that are referenced to the publication the sentence appears in.
A corpus of freely available journal papers available via PubMed consisting of about 10,000 publications has already been processed and the related arguments and metadata have been stored in triple stores that can now be queried. A subset of this corpus corresponding to the domain of cancer chemo prevention has also been publicly exposed.
Further Applications and Potential Usage
Argument and named entity extraction on unstructured text can be applied to any domain where published text tends to follow some predefined rules. Application in law, where sets of grammar rules have already been published can be straightforward. Newsfeeds, market reviews and political comments can also be addressed, especially if machine learning components are incorporated.



